

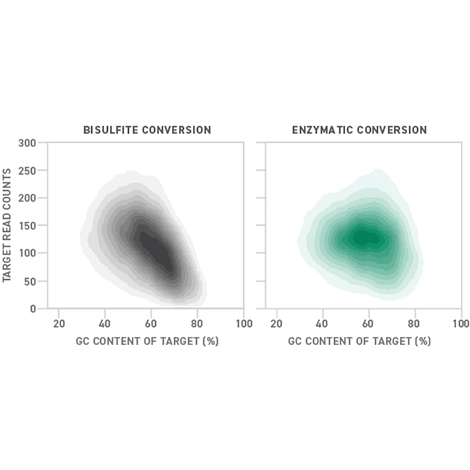
While most methods rely on bisulfite conversion of DNA to detect unmethylated cytosines, bisulfite treatment requires high temperatures and a harsh pH that can damage DNA, which is particularly challenging with small sample size or already fragile sample types.
With the Twist Methylation Detection System, you can rescue DNA molecules and experience uniform genomic coverage of more DNA regions by using an enzymatic conversion method. This method allows you to detect up to 15% more methylated cytosines, increasing confidence in methylated-base calls for the study of epigenetics.

Gahee Park, PhD, from the University of Cambridge shares how DNA methylation changes are robust cancer-specific markers, and how the Twist NGS Methylation Detection System helped her team improve enrichment accuracy three-fold, while decreasing experimental costs four-fold.
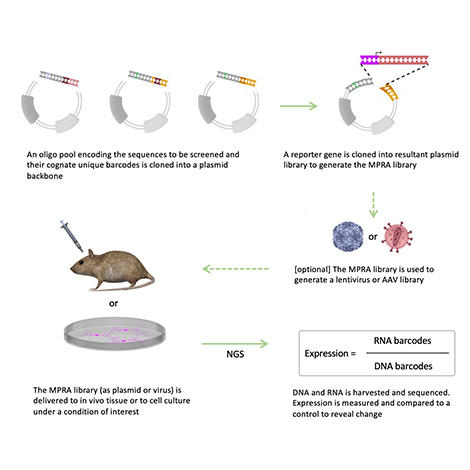
Without a detailed understanding of the elements that alter gene expression through epigenetic means, clinically relevant information coded within the growing influx of genetic data will remain undiscovered.
Researchers utilize Massively Parallel Reporter Assays (MPRAs) to interrogate regulatory elements in high throughput.
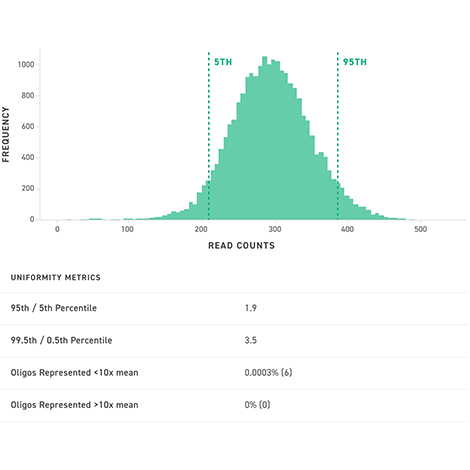
Our silicon platform allows us to quickly generate large, accurate and highly uniform MPRA libraries consisting of oligos up to 300 base pairs in length.
These libraries have many advantages:

DNA methylation is an epigenetic modification that can impact gene expression, and certain abnormal methylation patterns have shown correlation with cancer. While many researchers work toward developing tests to identify multiple cancers from a single blood sample, it remains complex. However, the Twist Targeted Methylation Detection System helps to meet those challenges. With its unique ability to rapidly generate immense numbers of oligonucleotide probes, Twist is able to simplify deconvolution and reduce bias while retaining high sensitivity to low-abundance DNA.
Epigenetics is the study of how the environment impacts gene expression through DNA modifications that do not alter the genome sequence. Instead, chemical modifications like methylation and acetylation impact several factors, including the folding of chromatin, the binding of histones, the activity of effectors, and the efficacy of promoters. Changes to the epigenome are linked to adverse health effects and are a known underlying risk factor for cancer and other complex diseases. The identification of new epigenetic hallmarks of disease, and the screening for known epigenetic markers, are vital to improving human health.
Want to learn more about Twist's innovative solutions for methyl-seq? Check out the e-book "Methylation Sequencing in Cancer Detection."

A complete methyl-seq workflow including enzymatic conversion capturing up to 15% more methylated cytosines

Custom target enrichment panels for precise, uniform capture of regions of interest involved in epigenetics

Highly uniform libraries of long oligos powering massively parallel functional genomic assays for interrogating gene expression
